Mechanized Process to Analyze Images Precisely
Scientists who have been seeking ways to stimulate growth of neurons and spending time in carefully analysing microscopic images of cells growing in petri dishes can now get help from a new procedure which has been developed by Brown University researchers.
They have mechanized the process to analyse images much more precisely than earlier automated methods.The initial round of testing and the method have been described in Nature’s Scientific Reports. As the neurons tend to grow, they seem to extend tough appendages known as neuritis which form grave connection with the nearby cells.
The networks of neurons and neurites are needed for healthy function of the nervous system. Scientists are concerned in discovering new ways of encouraging neuron growth through drugs, electrical stimulation or any other means. In order to test the effect of these attempts, they have grown neurons in the lab, with the application of various treatments to see if they could stimulate growth.
It generally involves taking pictures of hundreds of microscope of neurons as they tend to grow over a passage of hours or days.
Neurites –Tiny Structures – Difficult to View Under Microscope
Tayhas Palmore, professor of engineering at Brown as well as the senior author of the new paper, comments that `one is left with this massive stack of photos and the needs to analyse the changes from one image to the next could be really laborious’.
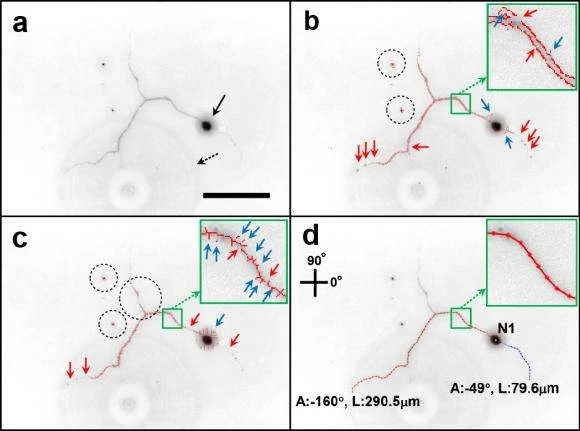
Details in those images seem to be dangerous. Neurites seems to be tiny structures which are difficult to be viewed under a microscope at the time of live-cell imaging. However, measuring their length and thickness precisely is essential in assessing stimulated cell growth.
A few methods are available which automate the image analysis, though they do not perform well. They tend to work by looking at individual pixels in an image, applying a uniform filter which tends to pick out pixels with the highest intensity and those high intensity pixels are presumed to be neuron and neurite structures.
The issue is that microscope images generally are not of high quality, thus making it difficult to discern cells structures from random artifacts which may be present in the image. This result in the filters often tends to include pixels which are not relevant to neuron structures, editing pixels which are important.
New Method – Neuron Image Analyzer – NIA
This seems to be a problem especially in measuring the tiny neurite appendages and the filters regularly fail to measure the complete extent of the neurite growth. A better solution was sought by a former graduate student in Patmore’s lab, Kwang-Min Kim who is now a postdoctoral researcher at Stanford.
Inspired by the earlier work of Kilho Son, a graduate student in computer vision as well as a co-first author on the paper, he created a new method which dispenses with the uniform filters utilised in other approaches. The new approach known as Neuron Image Analyzer – NIA takes into consideration how pixels tend to be related to the nearby pixels.
He had commented that they don’t just look for high intensity pixels but look at the relational information between pixels. This way they can trace pixels which are connected to each other that help in tracing the complete neuron structure. Another procedure adopted by the method is a particular statistical test which is good in picking out circular or elliptical structures which is utilised to locate precisely and measure the soma, the blob shaped main body of a neuron.